22. Exercise: K-means Estimator & Selecting K
Creating a
KMeans
Estimator
Now that we’ve run the original data through PCA and moved from 34-dimensional data to 7-dimensional component data, we’re better prepared to actually cluster this data! So, now, I’ll ask you to use these components that we’ve gotten from our training data and cluster counties using k-means.
You'll instantiate a
KMeans
estimator, by specifying specific model arguments and passing them into a KMeans constructor (
documentation, here
). Knowing how to read documentation is an important skill for learning to create models on your own!
Here is what this exercise looks like in the main, exercise notebook:
EXERCISE: Define a k-means model
Your task will be to instantiate a k-means model. A KMeans estimator requires a number of parameters to be instantiated, which allow us to specify the type of training instance to use, and the model hyperparameters.
# define a KMeans estimator
Some parameters in the KMeans documentation
General Estimator Parameters
From the documentation, you can see that you'll need to specify the IAM role (which we defined when creating the notebook instance), and details about the instance type to use for training.
Most of SageMaker's built-in algorithms are based off of an EstimatorBase object , which allows you to specify additional parameters. It is good practice to be specific about two additional parameters:
- output_path (str) – S3 location for saving the training result (model artifacts and output files). If not specified, results are stored to a default bucket.
- sagemaker_session (sagemaker.session.Session) – Session object which manages interactions with Amazon SageMaker APIs and any other AWS services needed.
Model-Specific Parameters
You'll also notice a parameter
k
for the number of clusters this model should produce as output. This parameter is specific to the k-means model, and for different models, you'll see different required model parameters.
Choosing a "Good" K
One method for choosing a "good"
k
, is to choose based on empirical data.
- A bad k would be one so high that only one or two very close data points are near it, and
- Another bad k would be one so low that data points are really far away from the centers.
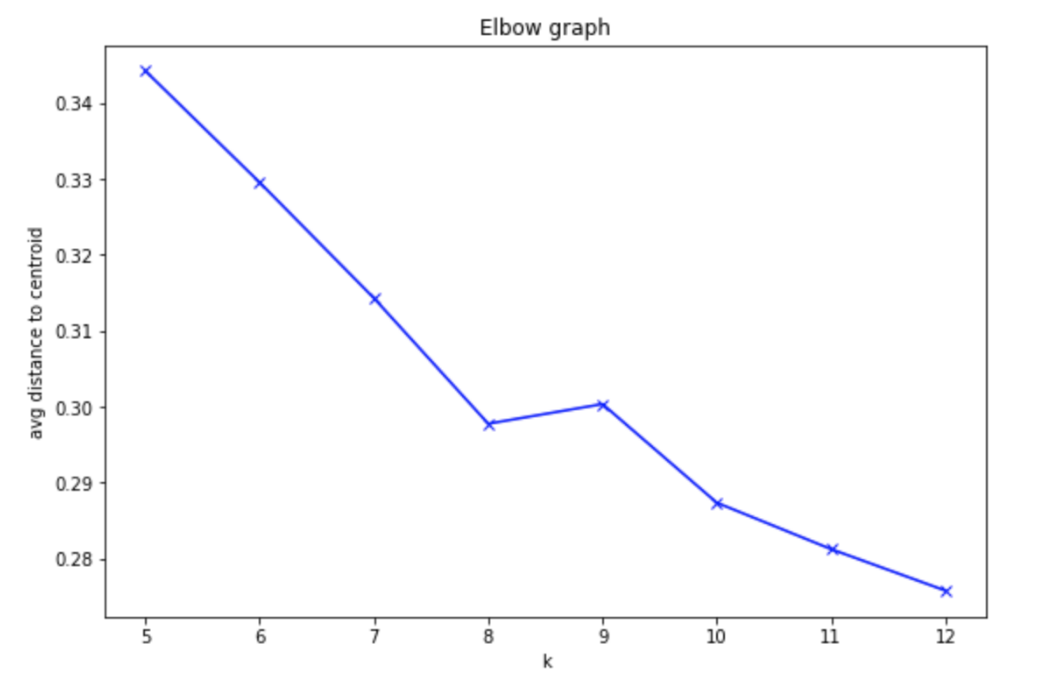
You want to select a k such that data points in a single cluster are close together but that there are enough clusters to effectively separate the data. You can approximate this separation by measuring how close your data points are to each cluster center; the average centroid distance between cluster points and a centroid. After trying several values for k, the centroid distance typically reaches some "elbow"; it stops decreasing at a sharp rate and this indicates a good value of k.
The graph below indicates the average distance—between our component data and cluster centroids—for a value of k between 5 and 12.
Training Job
After creating a KMeans estimator, I also want you to proceed with creating a training job. You'll have to format your data correctly for this job and make sure you are passing in the reduced-dimensionality training data. It may be helpful to reference the PCA training job code.
Here is what these exercises look like in the exercise notebook:
EXERCISE: Create formatted, k-means training data
Just as before, you should convert the counties_transformed df into a numpy array and then into a RecordSet. This is the required format for passing training data into a KMeans model.
# convert the transformed dataframe into record_set dataEXERCISE: Train the k-means model
Pass in the formatted training data and train the k-means model.
%%time
# train kmeansAfter you are done with these steps, you can move on to model deployment!